1 Introduction
2 GWAS Catalog with a ChIP-Seq Experiment
Here, we are gonna analyze the relation between transcription factor binding (ESRRA binding data) from a ChIP-Seq experiment and the genome-wide associations between DNA variants and phenotypes like diseases. For this task, we are gonna use a the gwascat package distributed by the EMBL (European Molecular Biology Laboratories).
First, we need to download the data, keep the 24 chromosomes (from 1 to Y) and, specify the sequence information from the GRCh38 human genome annotation.
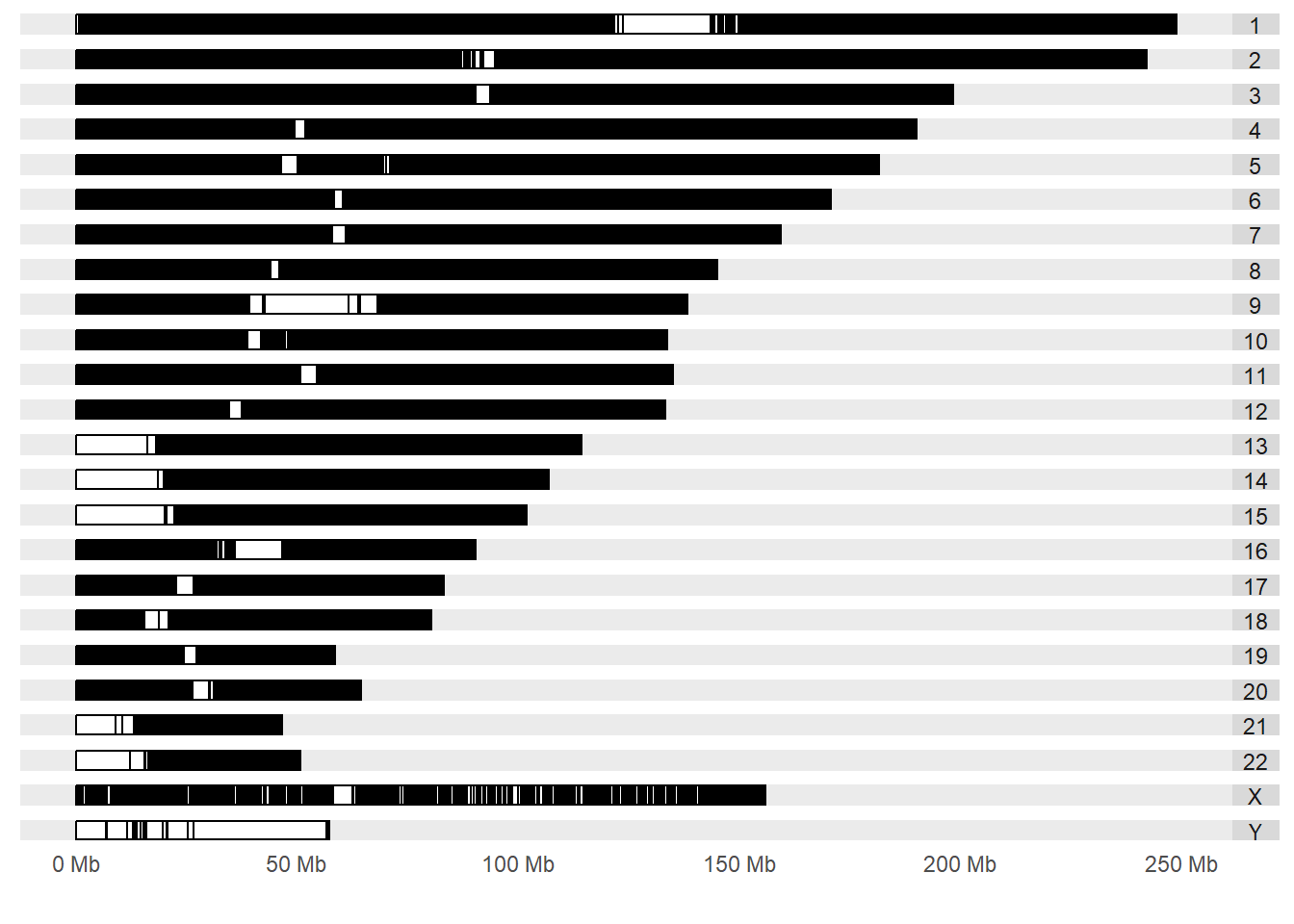
Now, let’s plot a karyogram that will show the SNP’s identified with significant associations with a phenotype. The SNP’s in the GWAS catalog have a stringent criterion of significance and there has been a replication of the finding from a independent population.
We can see the peak data as a GRanges object:
GRanges object with 1873 ranges and 7 metadata columns:
seqnames ranges strand | name score col
<Rle> <IRanges> <Rle> | <numeric> <integer> <logical>
[1] chrX 1509355-1512462 * | 5 0 <NA>
[2] chrX 26801422-26802448 * | 6 0 <NA>
[3] chr19 11694102-11695359 * | 1 0 <NA>
[4] chr19 4076893-4079276 * | 4 0 <NA>
[5] chr3 53288568-53290767 * | 9 0 <NA>
... ... ... ... . ... ... ...
[1869] chr19 11201120-11203985 * | 8701 0 <NA>
[1870] chr19 2234920-2237370 * | 990 0 <NA>
[1871] chr1 94311336-94313543 * | 4035 0 <NA>
[1872] chr19 45690614-45691210 * | 10688 0 <NA>
[1873] chr19 6110100-6111252 * | 2274 0 <NA>
signalValue pValue qValue peak
<numeric> <numeric> <numeric> <integer>
[1] 157.92 310.000 32 1991
[2] 147.38 310.000 32 387
[3] 99.71 311.660 32 861
[4] 84.74 310.000 32 1508
[5] 78.20 299.505 32 1772
... ... ... ... ...
[1869] 8.65 7.281 0.26576 2496
[1870] 8.65 26.258 1.99568 1478
[1871] 8.65 12.511 1.47237 1848
[1872] 8.65 6.205 0.00000 298
[1873] 8.65 17.356 2.01323 496
-------
seqinfo: 93 sequences (1 circular) from hg19 genomeIf we see the bottom of the GRanges table, this experiment have the hg19 annotation from the human genome. To work on the GRCh38 annotation we need to lift-over with a .chain file. For this we can use the AnnotationHub package.
AnnotationHub with 1 record
# snapshotDate(): 2023-10-23
# names(): AH14150
# $dataprovider: UCSC
# $species: Homo sapiens
# $rdataclass: ChainFile
# $rdatadateadded: 2014-12-15
# $title: hg19ToHg38.over.chain.gz
# $description: UCSC liftOver chain file from hg19 to hg38
# $taxonomyid: 9606
# $genome: hg19
# $sourcetype: Chain
# $sourceurl: http://hgdownload.cse.ucsc.edu/goldenpath/hg19/liftOver/hg19To...
# $sourcesize: NA
# $tags: c("liftOver", "chain", "UCSC", "genome", "homology")
# retrieve record with 'object[["AH14150"]]' We can find overlaps between the GWAS catalog and the ESRRA ChIP-Seq experiment but, there is a problem; the GWAS catalog is a collection of intervals that reports all significant SNPs and there can be duplications of SNPs associated to multiple phenotypes or the same SNP might be found for the same phenotype in different studies.
We can see the duplications with the reduce function from IRanges package:
[1] 261160We can see that there are 261160 duplicated loci. Let’s find the overlap between the reduced catalog and the ChIP-Seq experiment:
Hits object with 613 hits and 0 metadata columns:
queryHits subjectHits
<integer> <integer>
[1] 6 237757
[2] 9 221130
[3] 12 25060
[4] 15 104699
[5] 15 104700
... ... ...
[609] 1928 246305
[610] 1929 244698
[611] 1929 244699
[612] 1931 250380
[613] 1931 250381
-------
queryLength: 1932 / subjectLength: 268560We can see 613 hits. Then, we are gonna eobtain the ranges from those hits, retrieve the phenotypes (DISEASE/TRAIT) and show the top 20 most common phenotypes with association to SNPs that lies on the ESRRA binding peaks.
Distinct phenotypes identified on the peaks:
[1] 613Now, how to do the inference of these phenotype on peaks of these b cells? We can use permutation on the genomic positions to test if the number of phenotypes found is due to chance or not.
[1] 03 Explore the TCGA
Please check the dedicated script (top left) to see how to explore and get insights from the TCGA (The Cancer Genome Atlas) database.
4 Conclusion
References
Citation
@online{palacios_bernuy2024,
author = {Palacios Bernuy, Piero},
title = {Integrative {Analysis} of {Multi-omic} {Data}},
date = {2024-05-01},
langid = {en},
abstract = {This document is part of a series of the analysis of Omics
data. Especifically, here is showed how to analyze bulk RNA-Seq data
with Bioconductor packages. Also, it’s showcased how to make plots
of the RNA data in the context of differentially gene expression and
gene-sets.}
}