1 Methylation Analysis
1.1 Methylation
Epigenetics is the study of how your behaviors and environment can cause changes that affect the way your genes work. Unlike genetic changes, epigenetic changes are reversible and do not change your DNA sequence, but they can change how your body reads a DNA sequence (CDC,2024).
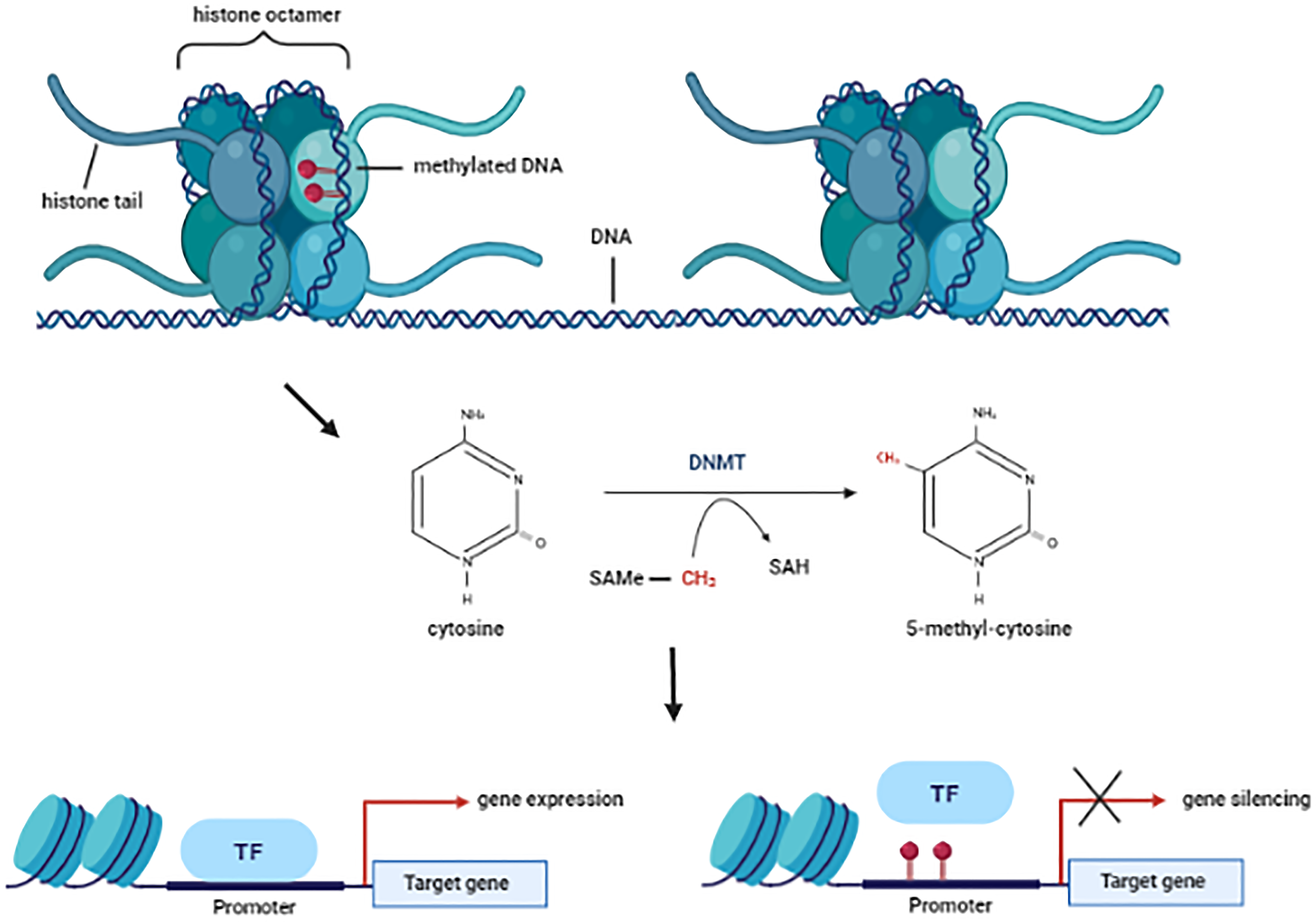
Talking about the genes, these can only be expressed when the chromatin is in it’s relaxed form (euchromatin) and reversely, these cannot be expressed in it’s compacted form (hetero-chromatin). Methylation of cytosine’s control part of this process of the chromatin and it’s really important to understand is a gene (or the surroundings) is methylated.
1.2 CpG’s
The cytosine’s (C) that are methylated are followed by a guanine (G). This pattern is called CpG in the context of epigenetics. These CpG’s tend to cluster into groups (CpG islands) and, these Cpg islands tend to be close to the promoter region of the genes.
1.3 Bisulfite treatment
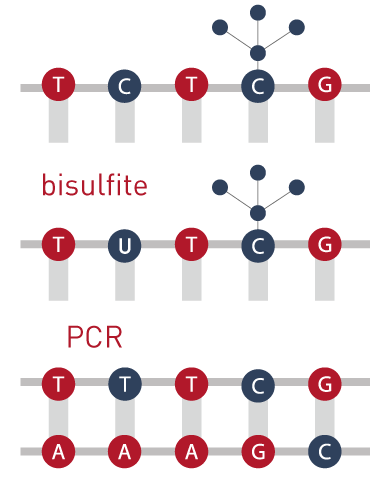
If a C is methylated the bisulfite treatment won´t change the cytosine but, if the C is not methylated, the C will suffer a transition to a T.
2 Methyl-Seq
Analyzing bisulfite sequencing data requires a server or an HPC. U can use the methylseq pipeline from nfcore to get the .cov files to proceed with the downstream analysis on R.
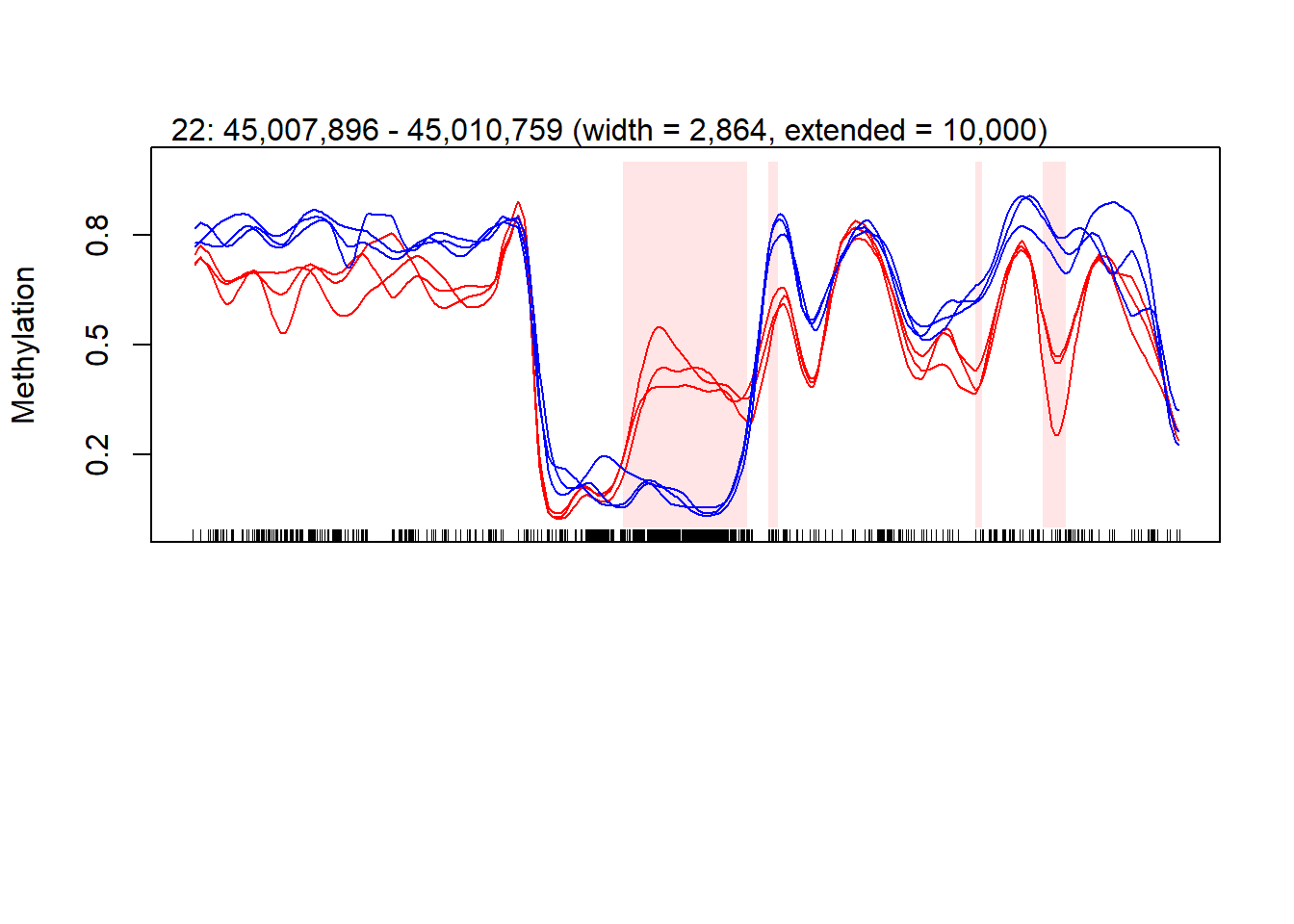
To see how to analyze Whole-genome bisulfite sequencing (WGBS) with the (Hansen, Langmead, and Irizarry 2012), click on the script below the image.
2.0.1 WGBS
3 Illumina 450k array
3.1 Methylation 450k array
To see how to analyze from a Illumina 450k array with the (Fortin, Triche, and Hansen 2016) package, click to the script below the image.
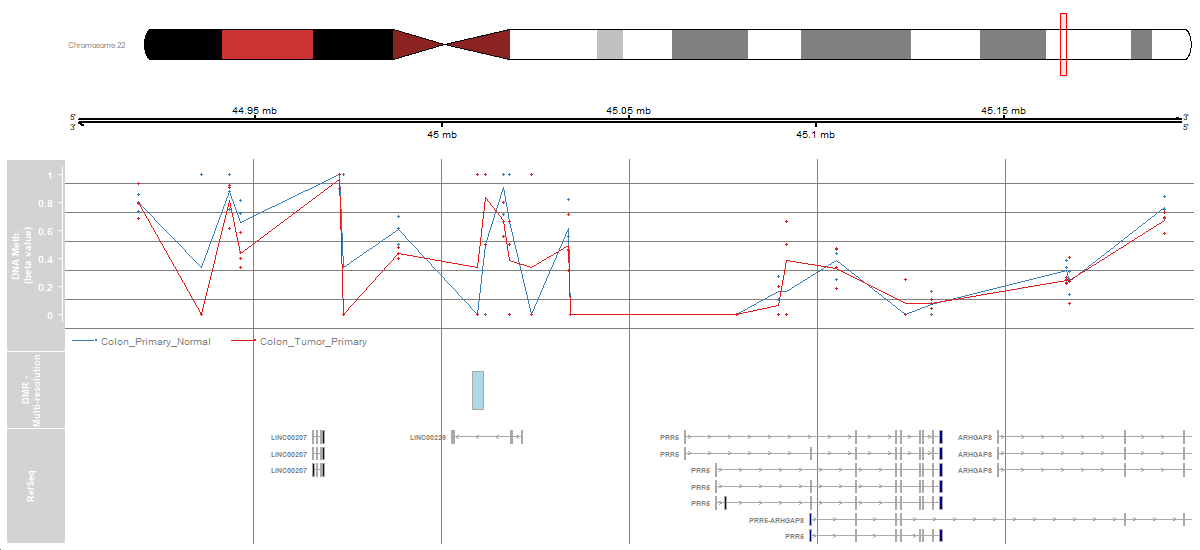
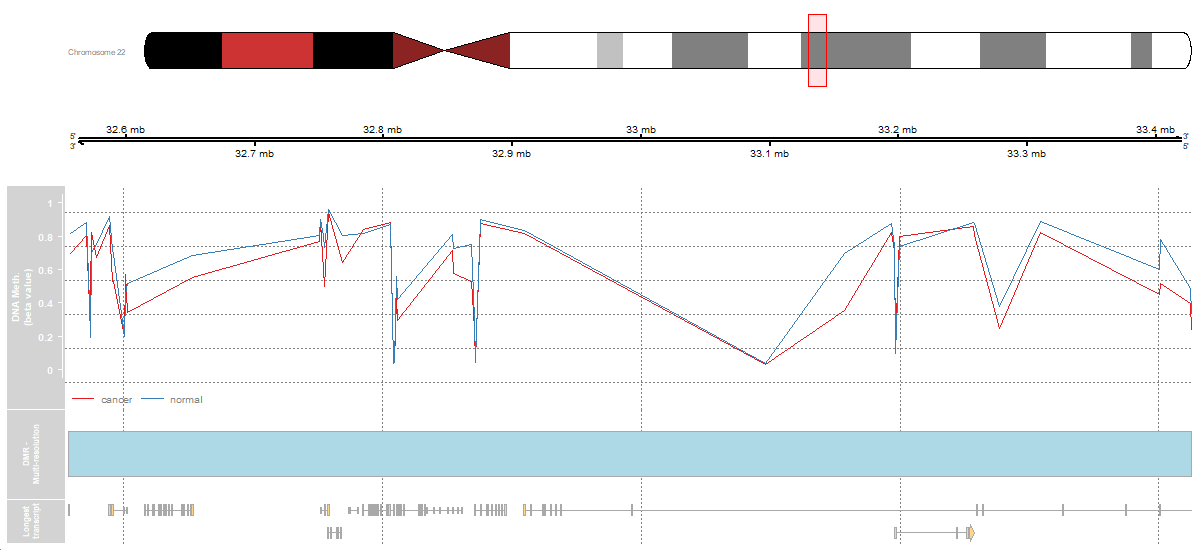
3.2 Methylation 450k array - Multi-resolution Analysis
To see how to analyze at a multi-resolution level from a Illumina 450k array with the (Fortin, Triche, and Hansen 2016) package, click to the script below the table.
Citation
@online{palacios bernuy2024,
author = {Palacios Bernuy, Piero},
title = {Analysis of {Methylated} {DNA}},
date = {2024-03-07},
langid = {en},
abstract = {This document is part of a series of the analysis of Omics
data. Especifycally, here is showed how to analyze Illumina 450k
array and Methyl-Seq data with Bioconductor packages. Also, it’s
showcased how to make plots of the methylation data in the context
of genomic positions and genes strutctures.}
}